Use #2: Out of Pipeline
If you do not have access to the raw data used in our article, you can still apply the key demcrit functions to your own, similarly structured dataset. In this vignette, we show how to do that using a small reproducible example.
Before you start, make sure you have a local installation of the demcrit R package. If not, see the package overview for installation instructions.
Setup
Start by loading the required packages:
Data Simulation
Because the original dataset cannot be shared, we will simulate a similar dataset using built-in functions:
set.seed(87542) # for reproducibility
data <- simulate_pdd_data()You can inspect the default parameters used in this simulation via:
prepare_defaults()
#> $Mu
#> FAQ MMSE MoCA sMoCA
#> 4.05 26.69 24.07 11.26
#>
#> $Sigma
#> FAQ MMSE MoCA sMoCA
#> FAQ 23.912100 -2.279718 -4.594644 -3.483636
#> MMSE -2.279718 4.928400 4.867128 3.284712
#> MoCA -4.594644 4.867128 12.110400 9.058440
#> sMoCA -3.483636 3.284712 9.058440 7.507600
#>
#> $cens
#> [,1] [,2]
#> FAQ 0 30
#> MMSE 0 30
#> MoCA 0 30
#> sMoCA 0 16
#>
#> $crits
#> IADL IADL_thres cognition cognition_thres
#> MMSE (1) FAQ 7 MMSE 26
#> MMSE (2) FAQ9 1 MMSE 26
#> MoCA (1) FAQ 7 MoCA 26
#> MoCA (2) FAQ9 1 MoCA 26
#> sMoCA (1) FAQ 7 sMoCA 13
#> sMoCA (2) FAQ9 1 sMoCA 13This produces the following example dataset:
#> # A tibble: 1,218 × 8
#> id FAQ MMSE MoCA sMoCA FAQ9 type PDD
#> <glue> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <lgl>
#> 1 s1 6 29 25 12 2 MMSE (1) FALSE
#> 2 s1 6 29 25 12 2 MMSE (2) FALSE
#> 3 s1 6 29 25 12 2 MoCA (1) FALSE
#> 4 s1 6 29 25 12 2 MoCA (2) TRUE
#> 5 s1 6 29 25 12 2 sMoCA (1) FALSE
#> 6 s1 6 29 25 12 2 sMoCA (2) TRUE
#> 7 s2 12 24 21 9 3 MMSE (1) TRUE
#> 8 s2 12 24 21 9 3 MMSE (2) TRUE
#> 9 s2 12 24 21 9 3 MoCA (1) TRUE
#> 10 s2 12 24 21 9 3 MoCA (2) TRUE
#> # ℹ 1,208 more rowsKey variables include:
-
id– patient identifier -
type– diagnostic algorithm used -
PDD– diagnosis result for each patient–algorithm pair
For more details about how the data are generated, see:
Dementia Rates
To compute dementia (PDD) rates, we first prepare a specification of diagnostic algorithms and variable mappings. Then, we pass these to the summarise_rates() function:
# Extract default parameters:
pars <- prepare_defaults()
# Define algorithm specifications:
algos <- with(pars, {data.frame(
type = rownames(crits),
glob = crits$cognition, glob_t = crits$cognition_thres,
iadl = crits$IADL, iadl_t = crits$IADL_thres,
atte = "", atte_t = NA,
exec = "", exec_t = NA,
memo = "", memo_t = NA,
cons = "", cons_t = NA,
lang = "", lang_t = NA
)})
# Define variable names:
vars <- data.frame(
name = c(unique(algos$glob), unique(algos$iadl)),
label = c(unique(algos$glob), unique(algos$iadl)),
type = "continuous"
)
# Compute PDD rates:
rates <- summarise_rates(
list(PDD = data, algorithms = algos),
vars = vars,
plot = FALSE # Setting plot to FALSE bacause it is calibrated the data from the study..
)
# Format results:
tab1 <- rates$table |>
select(type, Global, IADL, N, Rate) |>
gt_apa_table() |>
cols_label(
type ~ "Algorithm",
Global ~ "Cognitive deficit",
IADL ~ "IADL deficit"
)| Algorithm | Cognitive deficit | IADL deficit | N | Rate |
|---|---|---|---|---|
| MMSE (1) | MMSE < 26 | FAQ > 7 | 203 | 17 (8.37%) |
| MMSE (2) | MMSE < 26 | FAQ9 > 1 | 203 | 48 (23.65%) |
| MoCA (1) | MoCA < 26 | FAQ > 7 | 203 | 29 (14.29%) |
| MoCA (2) | MoCA < 26 | FAQ9 > 1 | 203 | 85 (41.87%) |
| sMoCA (1) | sMoCA < 13 | FAQ > 7 | 203 | 31 (15.27%) |
| sMoCA (2) | sMoCA < 13 | FAQ9 > 1 | 203 | 91 (44.83%) |
Pairwise Concordance
Next, we can compute pairwise concordance statistics across algorithms:
concs <- describe_concordance(list(PDD = data, algorithms = algos))For details about the resulting object, see:
We can visualize concordance matrices for several metrics:
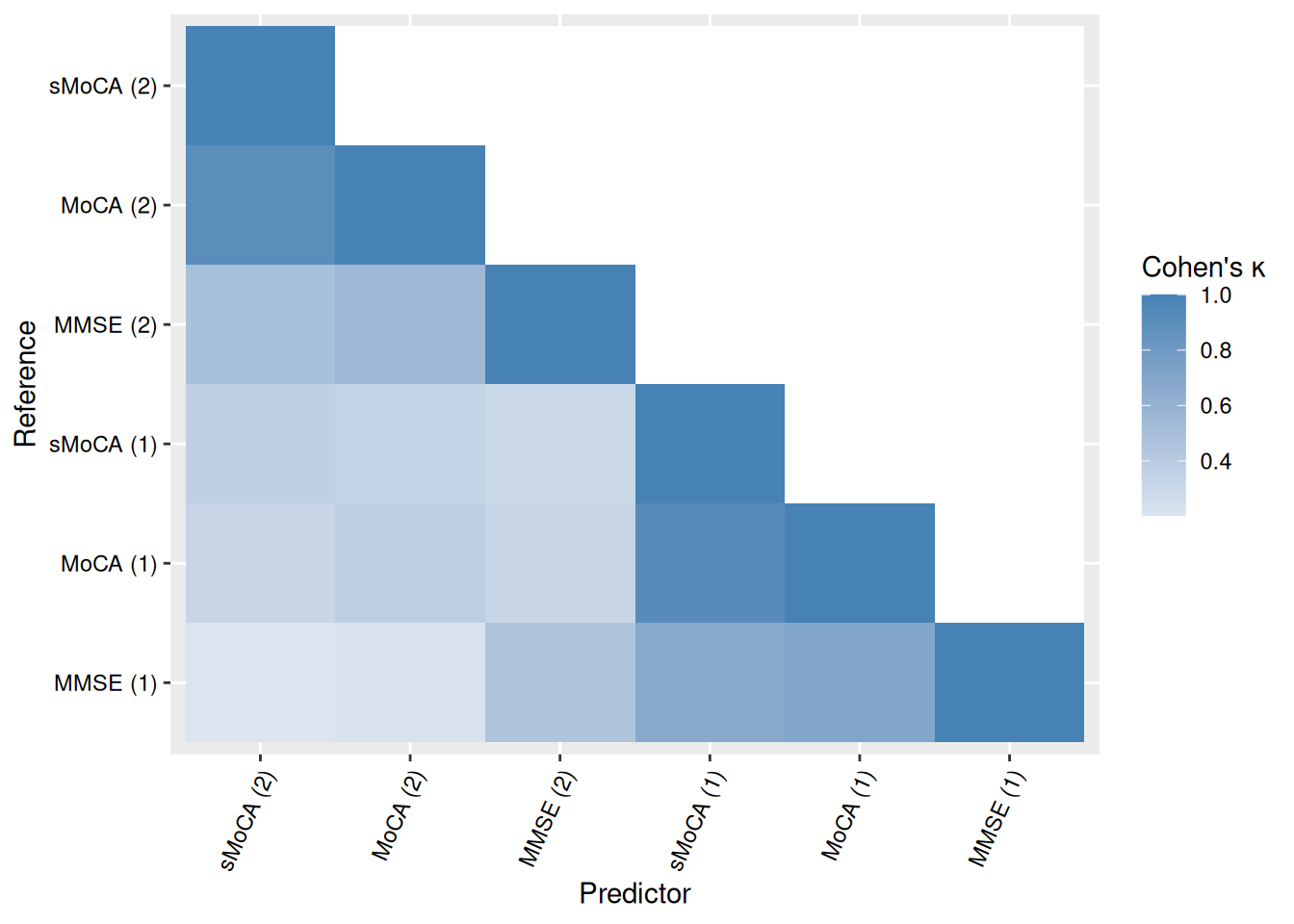
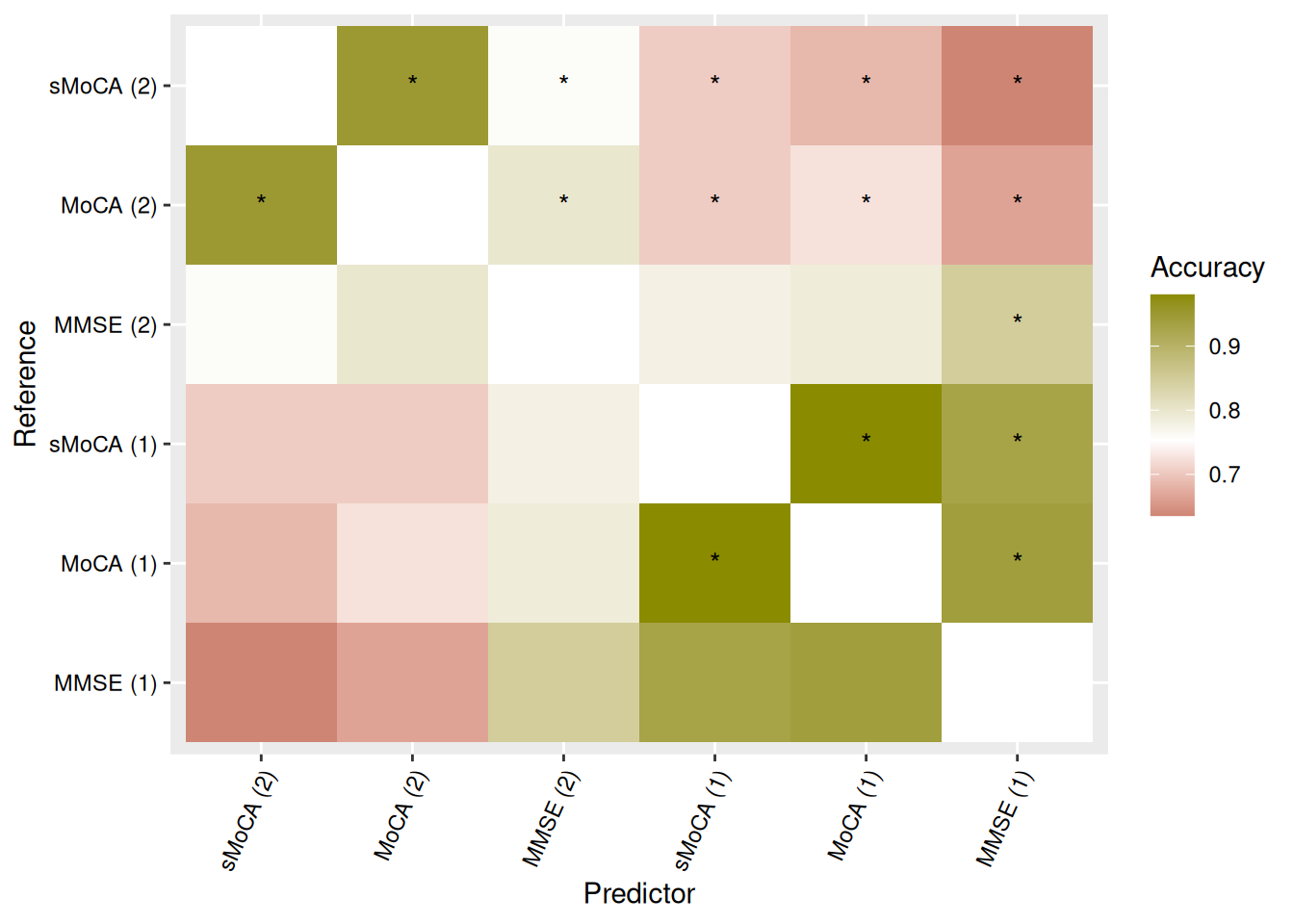
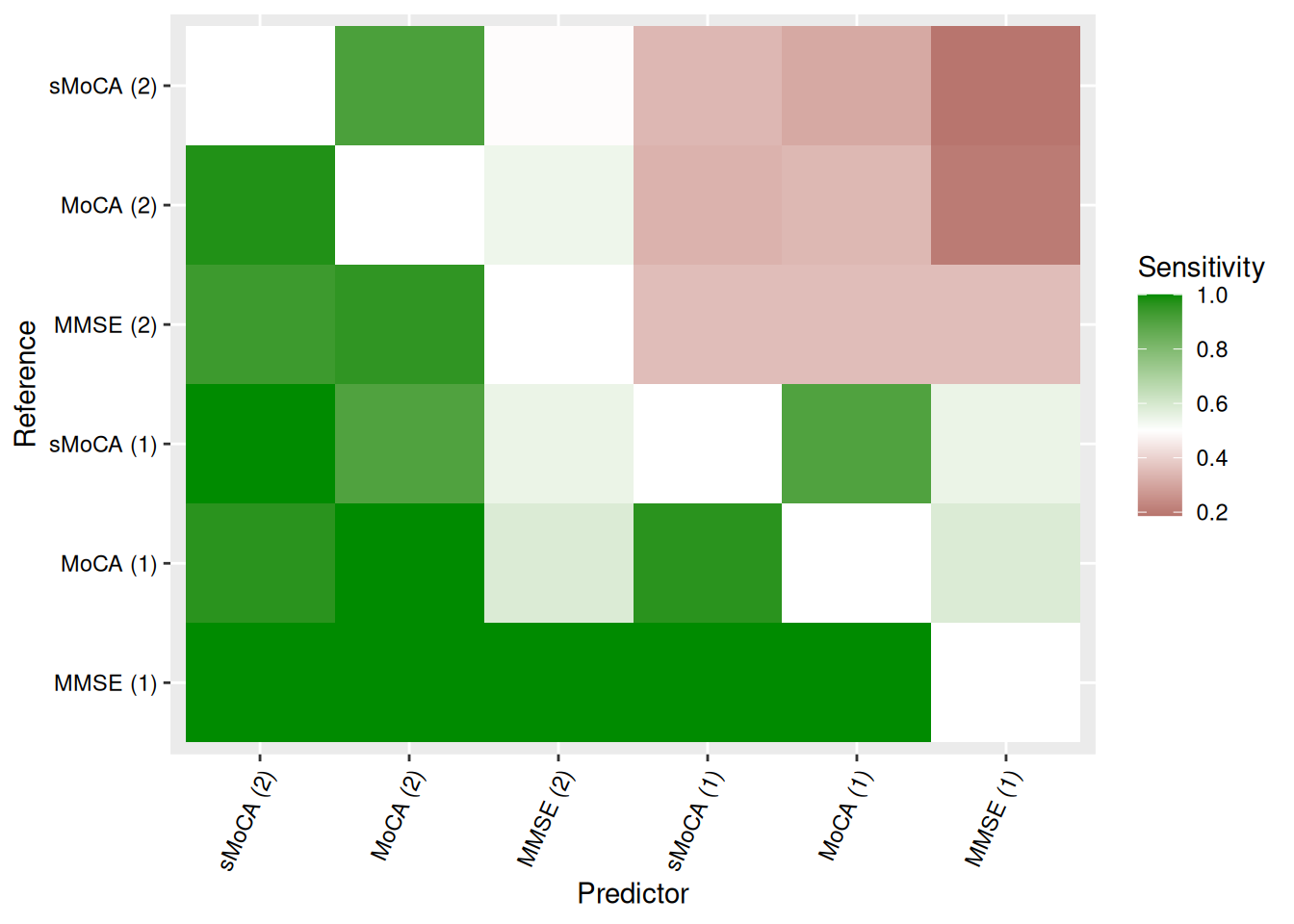
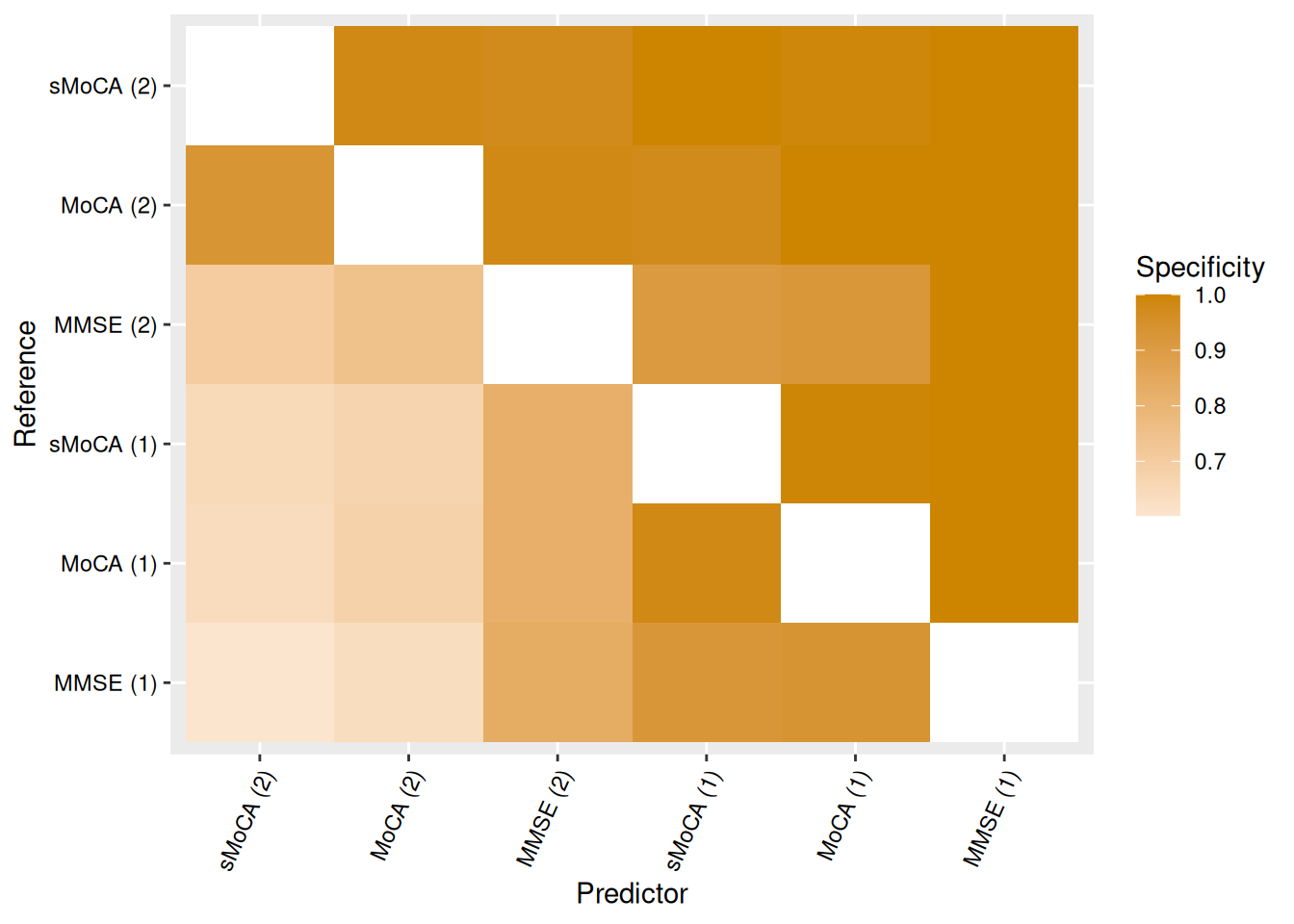
Alternatively, we can extract specific concordance statistics and present them in a table. Table Table 2 shows one such example, with algorithms sorted by raw accuracy in predicting MoCA (1).
tab2 <- concs$table |>
filter(reference == "MoCA (1)") |>
filter(reference != predictor) |>
arrange(desc(Accuracy_raw)) |>
select(predictor, Kappa, Accuracy, NoInformationRate_raw, AccuracyPValue) |>
mutate(AccuracyPValue = do_summary(AccuracyPValue, 3, "p")) |>
gt_apa_table() |>
fmt_number(decimals = 2) |>
cols_label(
predictor ~ "Predictor",
NoInformationRate_raw ~ "NIR",
AccuracyPValue ~ "p-value"
)| Predictor | Kappa | Accuracy | NIR | p-value |
|---|---|---|---|---|
| sMoCA (1) | 0.92 [0.85, 1.00] | 0.98 [0.95, 0.99] | 0.86 | < .001 |
| MMSE (1) | 0.71 [0.56, 0.86] | 0.94 [0.90, 0.97] | 0.86 | < .001 |
| MMSE (2) | 0.32 [0.17, 0.47] | 0.79 [0.73, 0.84] | 0.86 | .997 |
| MoCA (2) | 0.38 [0.27, 0.48] | 0.72 [0.66, 0.78] | 0.86 | 1.000 |
| sMoCA (2) | 0.32 [0.22, 0.42] | 0.68 [0.62, 0.75] | 0.86 | 1.000 |